| |
The first release of ABS provides two applications to aid during the automatical training of
programs that discover patterns in related sequences: CONSTRUCTOR and EVALUATOR.
In the DATABASE SERVICES menu, these programs are accessible through the options 2.4 and 2.5.
The construction of benchmarks
Constructor is a tool to produce artificial DNA sequences in which a subset of the binding sites
from ABS are inserted. The users can customize the content of the background sequence, the number
of motifs that are planted, the subset of the real sites that can be used, the density
of motifs on each sequence, and the length and the number of the sequences.
As a result, the generated sequences and the positions in which a subset of the motifs in ABS are displayed.
A graphical representation in which every line corresponds to a sequence and every box to motif is also
included at the bottom. For instance, here below you have 10 sequences in which 5 classes of motifs have
been inserted.
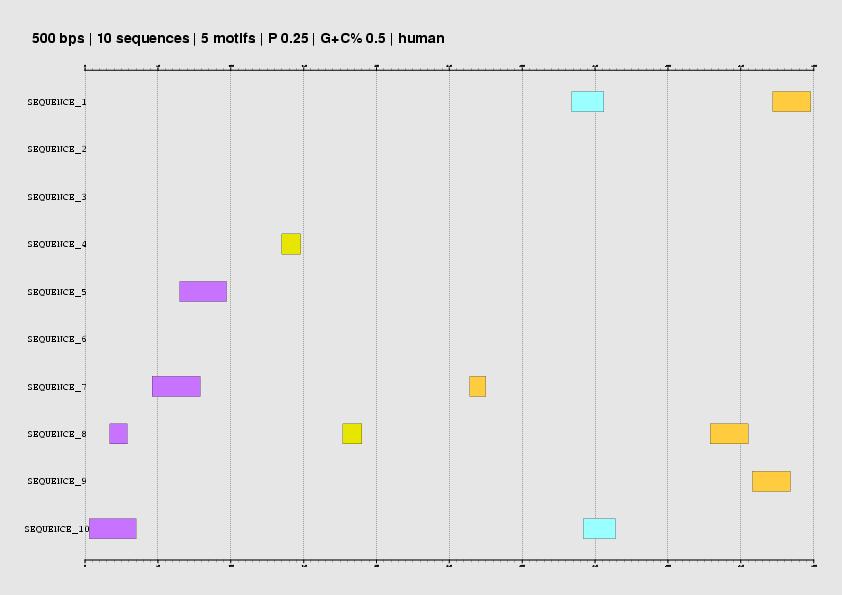
In theory, a pattern discovery program should recognize the conservation of at least the first grup of
binding sites of the same TF that are present in the sequences 5,7,8 and 10. The last motif is also
a good example to evaluate as it is present in the sequences 1,8 and 9.
To learn more about the creation of benchmarks go to the next webpage
The evaluation of benchmarks
The sequences created in the previous step can be input to a motif search program that will produce a list
of conserved motifs. Such motifs then must be compared to the correct coordinates of the real sites that
were inserted over there in order to measure the accuracy of such a program to predict conserved motifs in
related promoters (either coregulated genes or orthologous genes).
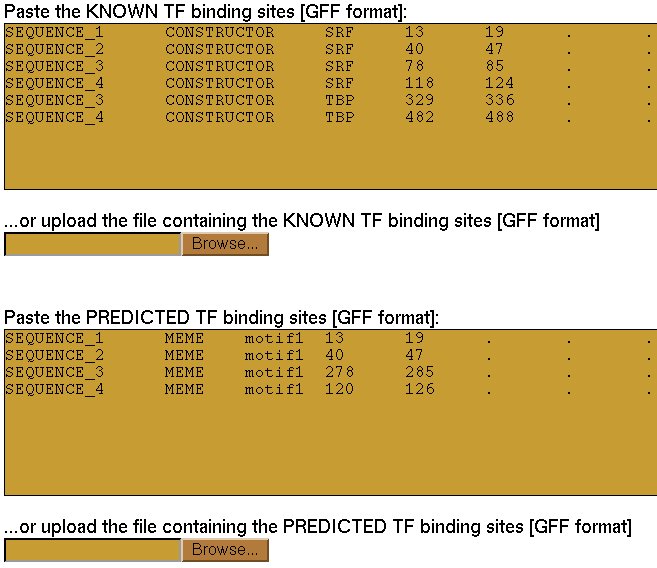
The program Evaluator uses the standard accuracy measures to assess the correctness of the predictions introduced
by the user in contrast to the real sites also submitted by the user. In the output, a table with the accuracy
at both nucleotide and site level is detailed. Moreover, the formal definitions of the values are always included
to facilitate the interpretation.
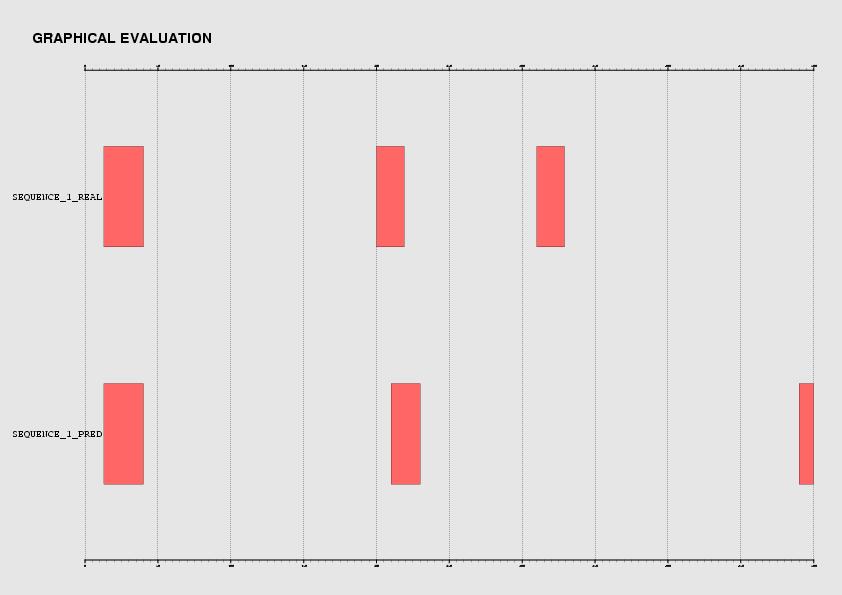
Furthermore, a graphical representation that associates real and predicted sites is provided:
To learn more about the evaluation of pattern discovery programs in ABS, follow this link
|
|