| |
This is a tutorial to show how the ABS database can be very useful in the process
of testing pattern discovery programs, and in the study of new forms of representation
for sets of binding sites associated to a given TF.
The ABS database is a collection of orthologous known binding sites with a rich information about
the promoters and the genes in which such sites have been experimentally defined. Moreover, ABS is
oriented to the study of the regulatory regions so that it is possible to access the binding sites and
the promoter sequences as well as to computational predictions obtained with popular position weight
matrices collections and promoter sequences global and local alignments.
The query server is based on a simple interface that permits the access to
different subsets of the data and the execution of PERL programs like Constructor and
Evaluator. Several cross-links are placed along the web to permit an easy communication
among differents data types. In addition, there is a documentation or help page associated
to major services that aims to aid the users to learn how to use the database.
We strongly recomend you to keep this tutorial in one window and use a secondary window
to browse along the database following the instructions. The main menu will be the starting
point for us at the address:
/datasets/abs2005/index.html
This is the screen you will be watching now:
The section 1 (PRESENTATION) is interesting for starting to understand what can be found here:

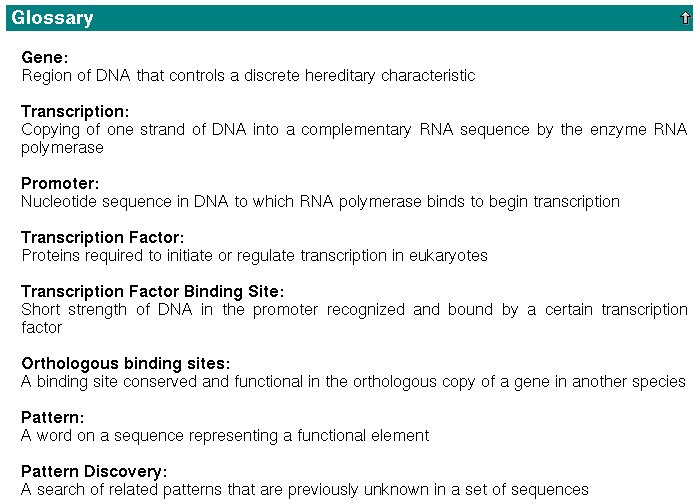
A short glossary can be even found here at the bottom of the page:
|
|