| |
The generation of artificial datasets to test the accuracy of motif search applications
is a very relevant aspect in the investigation of pattern discovery. The ABS database
provides a program Constructor to allow the users the creation and customization of their
own datasets for benchmarking tests.
We have decided to implement the generation with a system that first, chooses the real binding sites
from the ABS collections and second, embeds the sites in artificial sequences that are generated
according to the composition decided by the user.
The benchmark consists of N sequences that can be graphically represented horizontally as imaginary
lines in which the boxes represents the location of the motifs. For instance, this is a series of
examples showing the behaviour of the parameters (number of motifs and number of sequences):
The length of the sequences is easily modified by selecting one of the standard sizes (500,1000,2000,5000):
Moreover, the users can customize the probability that a motif is present or not in
the sequences (0..1), being able to produce more complex datasets like this:
Finally, it is possible to choose the composition of the background sequences and
the species from which the binding sites are coming from.
The output of Constructor is divided into 4 parts:
| I. The artificial sequences |

|
-> The embedded motifs are highlighted in uppercase.
|
| II. The positions of the real motifs in the artificial sequences |
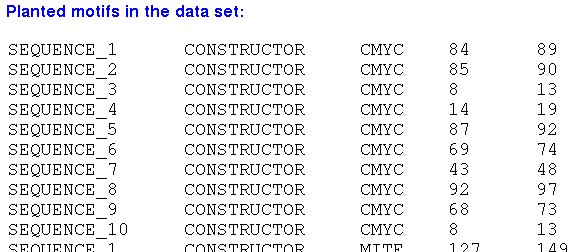
|
-> The annotations are shown in GFF format.
|
| III. The original annotations of these motifs in the ABS database |

|
-> The annotations are shown in GFF format.
|
IV. The graphical plot
|
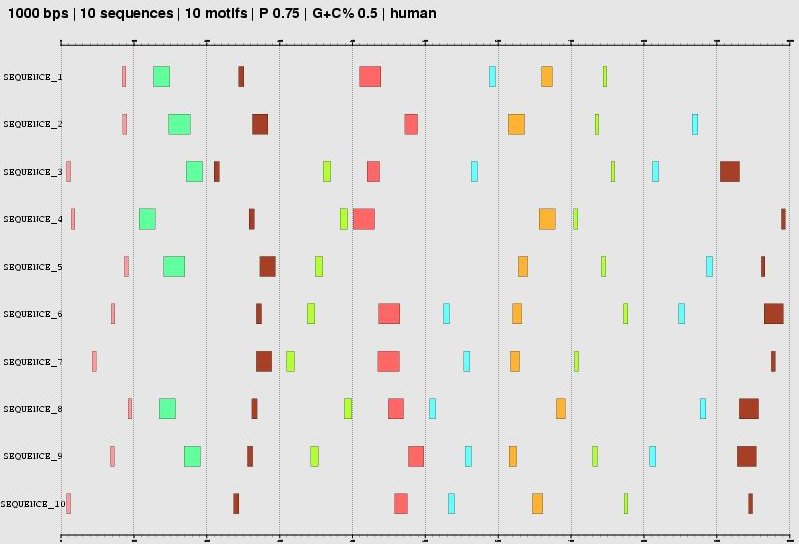
|
The complete example output can be examined here
CopyRight © 2005
ABS is under GNU General Public License.
|
|