|
|
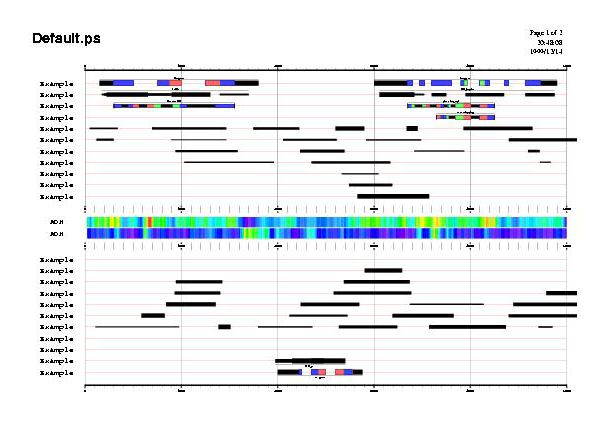
GFF2PS makes many assumptions on how things have to be displayed.
To illustrate this, the plot on the left was generated without any configuration file, only few command-line options, one of which forced two pages output, another one changed page size to "A5" format and the last defining a new title string.
Command-line was:
gff2ps -s a5 -P 2 -T "Default.ps" < GFF_sample.gff > Default.ps
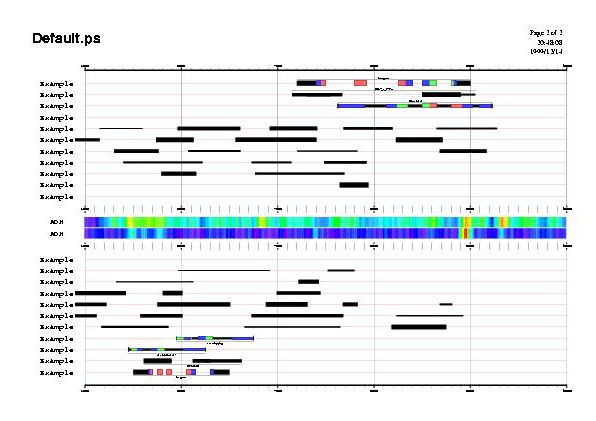
At top of the figures there is the header area, and from top to bottom, forward strand sources, the middle area for those features without defined strand, and reverse strand sources. Ordering for sources is taken from input files and is preserved on forward section but inverted on reverse section. In the middle area two scoring vectors are shown as color gradients.
The next three examples share the same GFF input file with current plot.
|