|
GFF2PS HOME PAGE |
Josep F. ABRIL's HOME PAGE |
| Back to Index |
| FILE/AUTHOR | PROGRAM | GFF record # | Strands | Group # | Strands | |||||
| + | . | - | + | . | - | |||||
| Adh | std1 | 513 | 246 | 0 | 267 | 43 | 20 | 0 | 23 | |
| std3 | 3756 | 1922 | 0 | 1834 | 221 | 117 | 0 | 104 | ||
| Vector | Fourier | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | |
| GCcontent | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | ||
| GROUP A | Birney | GeneWise | 252 | 148 | 0 | 104 | 184 | 111 | 0 | 73 |
| Borodovsky | GeneMarkHMM | 3219 | 1656 | 0 | 1563 | 265 | 140 | 0 | 125 | |
| Gaasterland | MAGPIE | 551 | 397 | 0 | 154 | 53 | 30 | 0 | 23 | |
| MAGPIE_exon | 1835 | 985 | 0 | 850 | 496 | 256 | 0 | 240 | ||
| Guigo | GeneID | 3342 | 1842 | 0 | 1500 | 1 | 0 | 0 | 1 | |
| Henikoff | BLOCKS | 346 | 193 | 0 | 153 | 0 | 0 | 0 | 0 | |
| Krogh | HMMGene | 2560 | 1389 | 0 | 1171 | 214 | 122 | 0 | 92 | |
| Mural | GRAIL | 855 | 477 | 0 | 378 | 21 | 0 | 0 | 21 | |
| Reese | Genie | 2358 | 1317 | 0 | 1041 | 0 | 0 | 0 | 0 | |
| GenieEST | 2547 | 1353 | 0 | 1194 | 0 | 0 | 0 | 0 | ||
| GenieESTHOM | 2694 | 1440 | 0 | 1254 | 0 | 0 | 0 | 0 | ||
| Solovyev | FGenesCGG1 | 3334 | 1668 | 0 | 1666 | 2 | 1 | 0 | 1 | |
| FGenesCGG2 | 598 | 324 | 0 | 274 | 133 | 58 | 0 | 75 | ||
| FGenesCGG3 | 8688 | 4341 | 0 | 4347 | 454 | 220 | 0 | 234 | ||
| GROUP B | Gaasterland | MAGPIE_Promoter | 421 | 220 | 0 | 201 | 0 | 0 | 0 | 0 |
| Ohler | LME_IMC | 3062 | 1508 | 0 | 1554 | 0 | 0 | 0 | 0 | |
| LME_SSM | 2552 | 1271 | 0 | 1281 | 0 | 0 | 0 | 0 | ||
| Reese | GeniePROM | 234 | 135 | 0 | 99 | 0 | 0 | 0 | 0 | |
| Werner | CoreInspectorTSS | 23 | 12 | 0 | 11 | 0 | 0 | 0 | 0 | |
| GROUP C | Benson | TandemRepeatFinder | 299 | 0 | 299 | 0 | 0 | 0 | 0 | 0 |
| Gaasterland | MAGPIE_Calypso | 1567 | 0 | 1567 | 0 | 0 | 0 | 0 | 0 | |
| TOTAL | 45608 | 22844 | 1868 | 20896 | 2089 | 1075 | 2 | 1012 | ||
| Back to Index |

|
| Josep, Moisés and the ISMB'99 Adh Poster. |
$BIN/gff2ps -VC ISMB_b0.rc -p -B 4 -P 3 -s b0 -- \
Adh.std1.gff Adh.std3.gff \
Birney.GeneWise.gff Borodovsky.GeneMarkHMM.gff Gaasterland.MAGPIE.gff \
Gaasterland.MAGPIE_exon.gff Guigo.GeneID.gff Henikoff.BLOCKS.gff \
Krogh.HMMGene.gff Mural.GRAIL.gff Reese.Genie.gff Reese.GenieEST.gff \
Reese.GenieESTHOM.gff Solovyev.FGenesCGG1.gff Solovyev.FGenesCGG2.gff \
Solovyev.FGenesCGG3.gff Gaasterland.MAGPIE_Promoter.gff Ohler.LME_IMC.gff \
Ohler.LME_SSM.gff Reese.GeniePROM.gff Werner.CoreInspectorTSS.gff \
Adh.Fourier.gff Benson.TandemRepeatFinder.gff Gaasterland.MAGPIE_Calypso.gff \
Adh.GCcontent.gff > ISMB1999_b0.ps 2> ISMB1999_b0.rpt
|
|
|
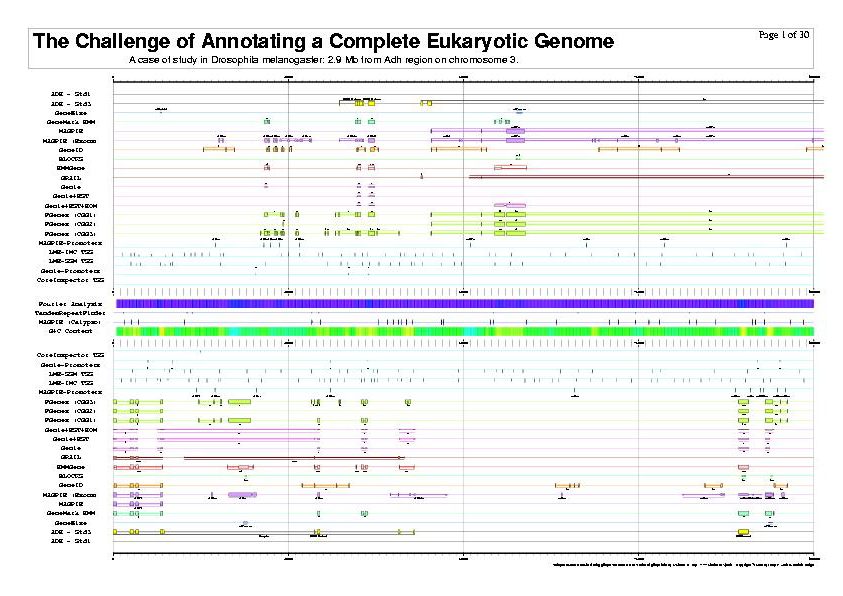
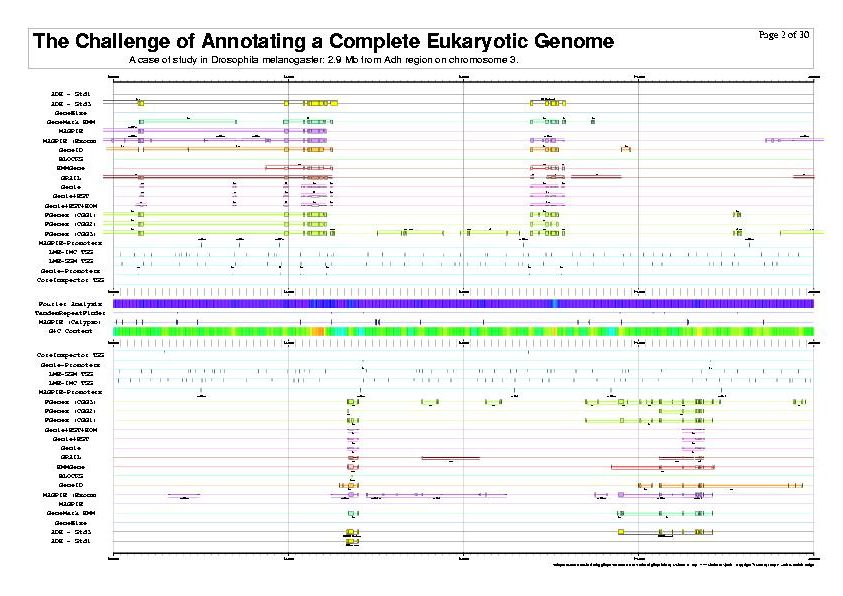
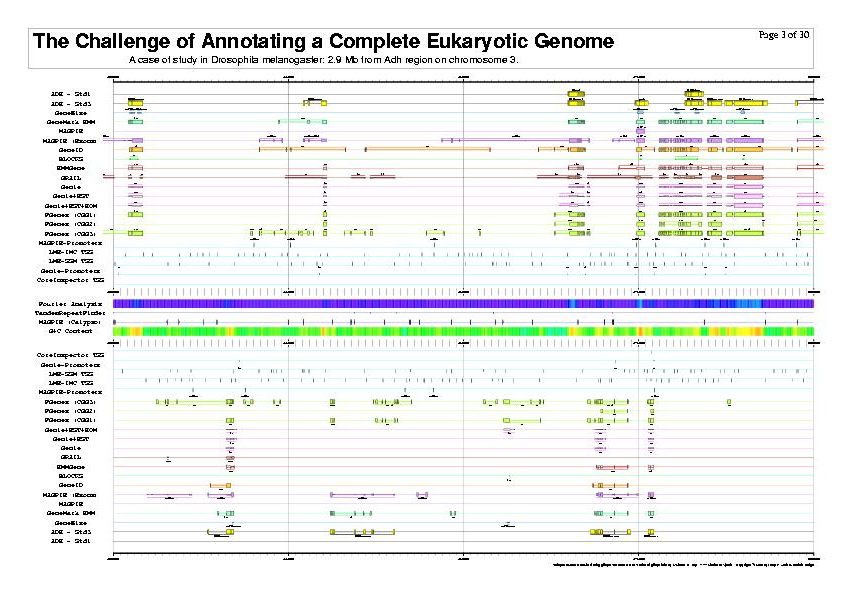
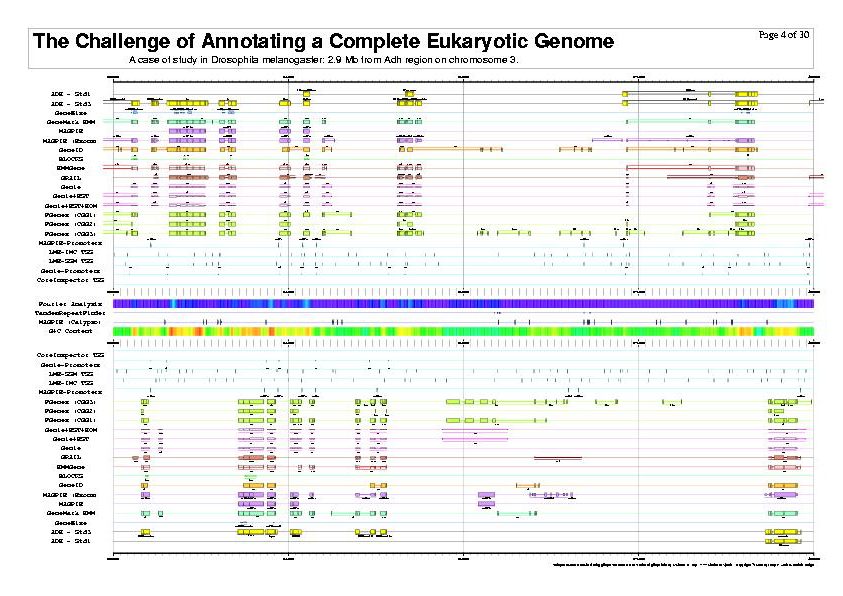
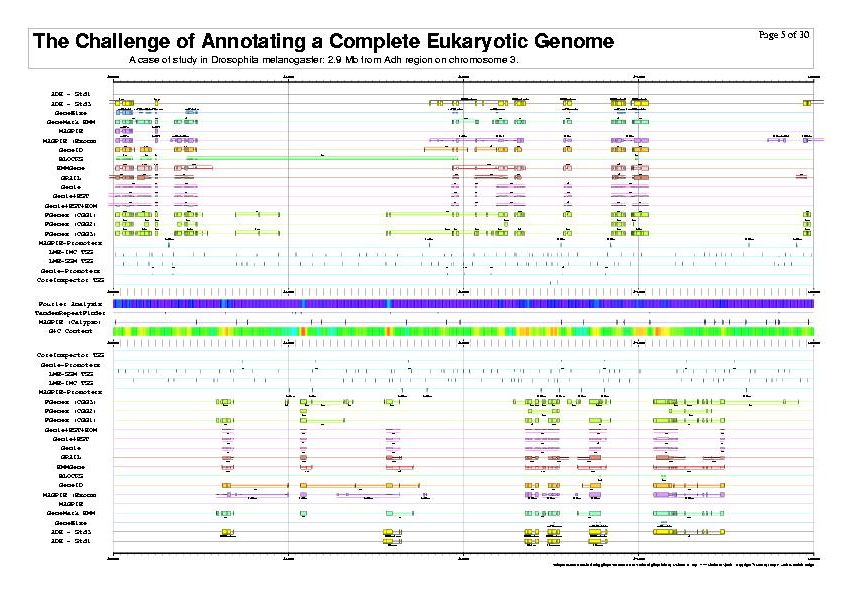
|
| The previous five figures are only the first five pages of a total of thirty pages. |
$BIN/gff2ps -VC ISMB_a4.rc -B 1 -N 100000 -s a4 -- \
Adh.std1.gff Adh.std3.gff \
Birney.GeneWise.gff Borodovsky.GeneMarkHMM.gff Gaasterland.MAGPIE.gff \
Gaasterland.MAGPIE_exon.gff Guigo.GeneID.gff Henikoff.BLOCKS.gff \
Krogh.HMMGene.gff Mural.GRAIL.gff Reese.Genie.gff Reese.GenieEST.gff \
Reese.GenieESTHOM.gff Solovyev.FGenesCGG1.gff Solovyev.FGenesCGG2.gff \
Solovyev.FGenesCGG3.gff Gaasterland.MAGPIE_Promoter.gff Ohler.LME_IMC.gff \
Ohler.LME_SSM.gff Reese.GeniePROM.gff Werner.CoreInspectorTSS.gff \
Adh.Fourier.gff Benson.TandemRepeatFinder.gff Gaasterland.MAGPIE_Calypso.gff \
Adh.GCcontent.gff > ISMB1999_a4.ps 2> ISMB1999_a4.rpt
|
|
|
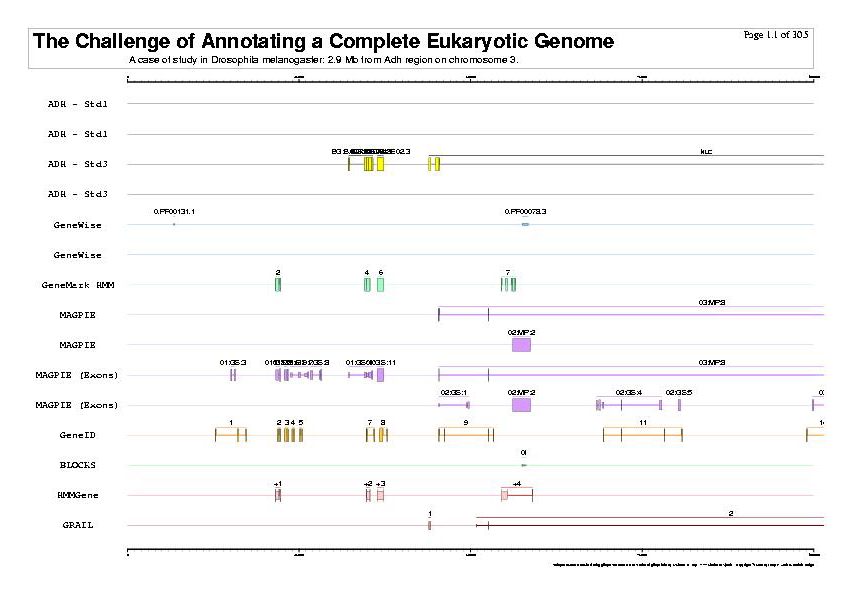
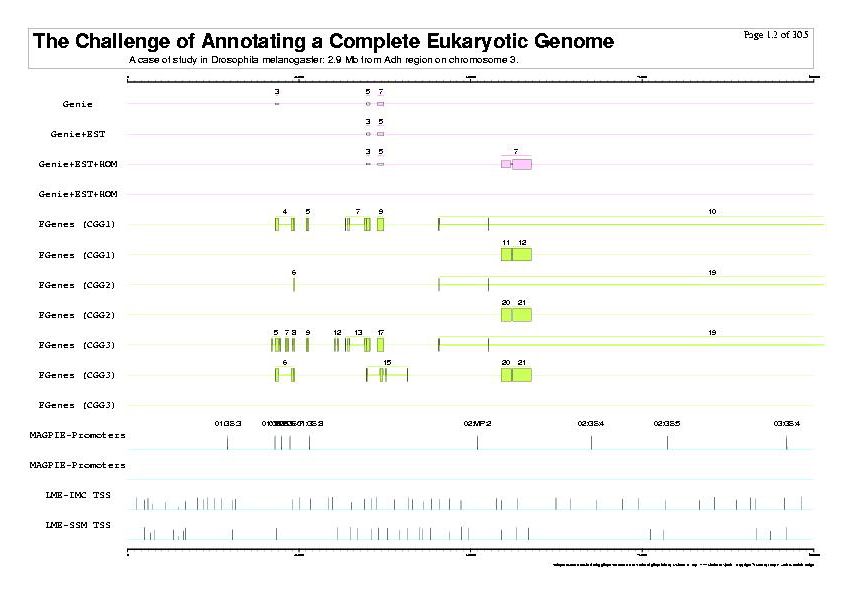
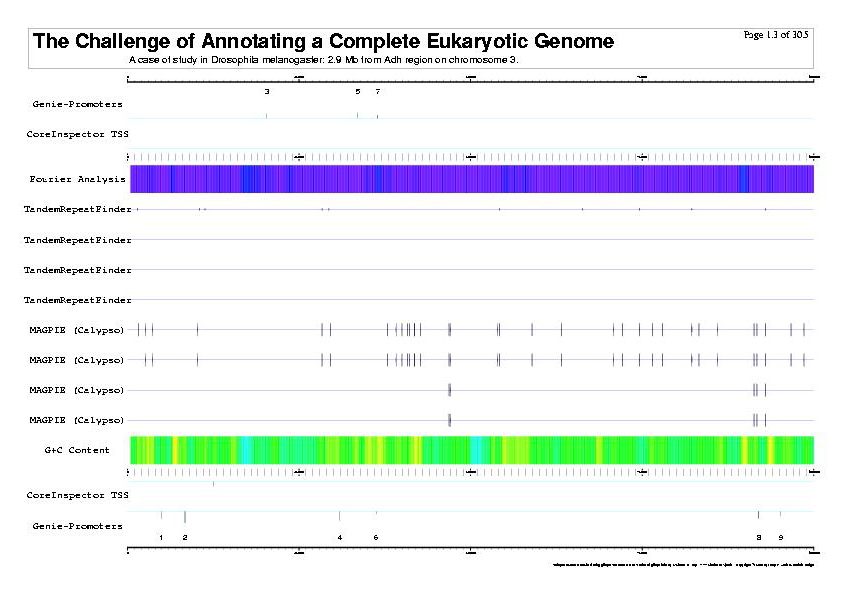
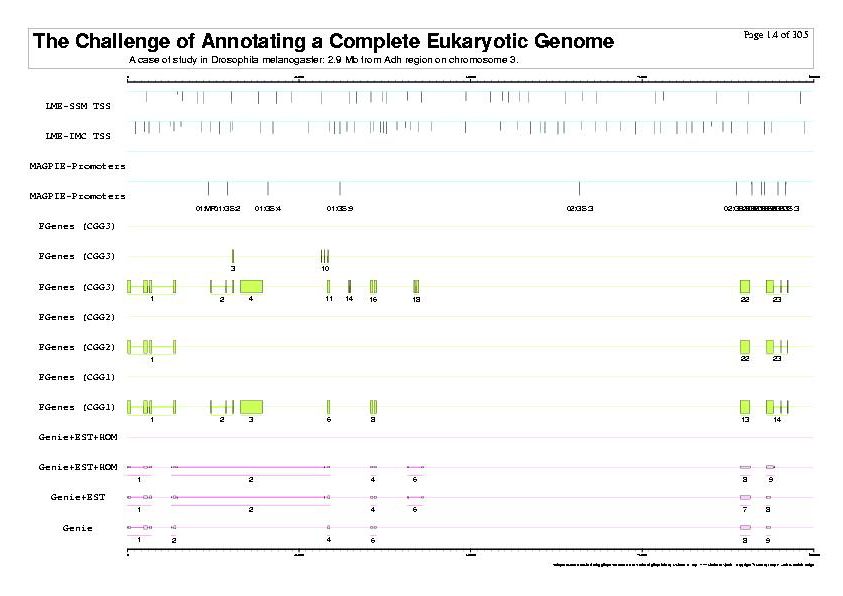
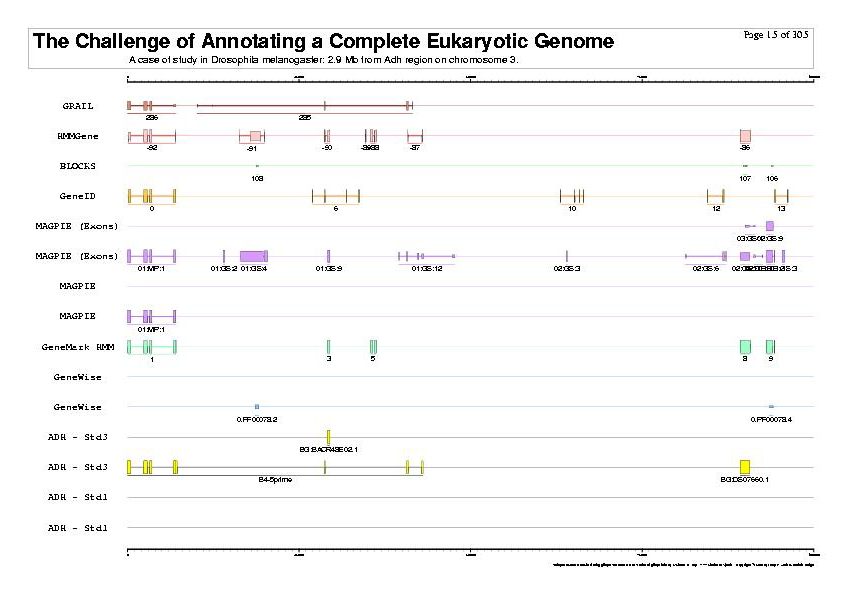
|
| The previous five figures are only the five vertical pages for the first horizontal page of a total of thirty by five pages. |
$BIN/gff2ps -VC ISMB_a4_VP.rc -B 0 -N 100000 -s a4 -- \
Adh.std1.gff Adh.std3.gff \
Birney.GeneWise.gff Borodovsky.GeneMarkHMM.gff Gaasterland.MAGPIE.gff \
Gaasterland.MAGPIE_exon.gff Guigo.GeneID.gff Henikoff.BLOCKS.gff \
Krogh.HMMGene.gff Mural.GRAIL.gff Reese.Genie.gff Reese.GenieEST.gff \
Reese.GenieESTHOM.gff Solovyev.FGenesCGG1.gff Solovyev.FGenesCGG2.gff \
Solovyev.FGenesCGG3.gff Gaasterland.MAGPIE_Promoter.gff Ohler.LME_IMC.gff \
Ohler.LME_SSM.gff Reese.GeniePROM.gff Werner.CoreInspectorTSS.gff \
Adh.Fourier.gff Benson.TandemRepeatFinder.gff Gaasterland.MAGPIE_Calypso.gff \
Adh.GCcontent.gff > ISMB1999_a4_VP.ps 2> ISMB1999_a4_VP.rpt
|
|
|
|
GFF2PS HOME PAGE |
Josep F. ABRIL's HOME PAGE |
| jabril@imim.es |