Selenoprofiles is a pipeline for profile-based protein finding in genomes.
Provided one or more protein alignments, it scans a target genome (or any other nucleotide database) and reports the gene structures of homologous genes. It can be used to easily characterize any protein family of interest across a massive amount of sequenced genomes, allowing a finely tuned filtering of results. Or it can be used with comprehensive sets of input profiles, in order to completely annotate by homology one or more genomes. The pipeline runs internally blast (psitblastn), exonerate (p2g mode) and genewise, combining them into a final set of non-overlapping predictions for all profiles.
Selenoprofiles is highly flexible. Even unexperienced user can edit the procedures of filtering, adapting them for each profile. The advanced user can plug-in its own code to customize the internal procedures of labelling, filtering, solving overlaps, outputing. It is also possible to write code to annotate genomic features of the predicted gene, such as protein or RNA motifs, which are stored and added to the native selenoprofiles output.
Although the program offers a variety of filtering methods, the default filter is quite effective. For each candidate, a measure of its similarity with the sequences in the profile is computed (AWSI score, see manual). The resulting score is compared to the distribution of this measure within the profile sequences. In this way, very conserved alignment profiles allow only highly similar sequences to pass the filter and be output.
Selenoprofiles can be used with any input protein family, but we initially developed it for selenoproteins. These peculiar proteins contain a selenocysteine, the 21st amino acid, which is inserted in correspondence to specific UGA codons, normally signalling translation termination. In selenoprotein transcripts we find specific secondary structures (SECIS elements), which targets a specific UGA to be read as Sec instead that as a stop. Since selenoproteins possess this peculiar feature (recoding of specific stop codons), normal gene prediction programs fail to predict them. Selenoprofiles in contrast can correctly predict selenoprotein genes, by using technical expedients to align selenocysteine positions. Selenoprofiles includes built-in profiles for selenoproteins and other proteins related to selenocysteine, allowing out-of-the-box prediction of these families.
Citation
Mariotti M, Guigó R. Selenoprofiles: profile-based scanning of eukaryotic genome sequences for selenoprotein genes. Bioinformatics. 2010 Nov 1;26(21):2656-63. Epub 2010 Sep 21
(Note that the paper refers to outdated version 1)
Installation
All "slave" programs run by selenoprofiles must be installed by user:
- Blast: ftp://ftp.ncbi.nlm.nih.gov/blast/executables/legacy/2.2.26/ [blastall legacy suite required (2.2.2x). Blast+ programs are not supported yet]
- Exonerate: https://www.ebi.ac.uk/about/vertebrate-genomics/software/exonerate
- Genewise: ftp://ftp.ebi.ac.uk/pub/software/unix/wise2/
- Mafft: https://mafft.cbrc.jp/alignment/software/
If you experience problems in the installation of these programs, this page may help you.
Selenoprofiles can be installed on any unix system with python 2.6 or newer. A python command line installer (install_selenoprofiles.py) is provided inside the installation package. You can get the installation package at https://github.com/marco-mariotti/selenoprofiles; simply open a terminal and type:
git clone https://github.com/marco-mariotti/selenoprofiles
cd selenoprofilesgit clone https://github.com/marco-mariotti/selenoprofiles
cd selenoprofilesAfter obtaining the package, follow the instructions on the README file included in the package. The simplest installation, suitable to be used with your own custom profiles, can be performed with:
python install_selenoprofiles.py -minpython install_selenoprofiles.py -minAlternatively, you can perform a full installation. This is necessary to scan with the built-in profiles for selenoproteins and Sec machinery. Selenoprofiles requires a few accesory files for this purpose, including the large Uniprot uniref50 database. If you need to use Selenoprofiles for this purpose, run:
python install_selenoprofiles.py -fullpython install_selenoprofiles.py -fullFor more information run:
python install_selenoprofiles.py --helppython install_selenoprofiles.py --helpThe manual is included within the installation package. However you can also download it here:
NOTE: Starting from version 3.4, due to the unstainable growth of the NCBI NR database, the full installation Selenoprofiles employs the Uniprot database of Uniref50 as protein reference for the methods tag score and GO score. This may cause slight differences in the performance of the built-in selenoprofiles and Sec machinery profiles when comparing with older versions.
Output and accessory programs
Several output formats are available, such as gff or fasta for nucleotide or protein sequences. The manual explains how to activate the built-in output types, and how to customize output by adding information of interesdt. By default, selenoprofiles produce only two type of output files: a fasta alignment of all predictions aligned to their corresponding profile, and a human readable p2g file showing the alignment gene structure and sequence (find an example of p2g file in here). The selenoprofiles package then contains a few additional programs, suited for projects aimed at searching certain protein families in many target species. The program selenoprofiles_join_alignments retrieves and merge into a single alignment all the results in different species. Then,* selenoprofiles_tree_drawer* allows their visualization in the phylogenetic tree of the target species. This program require the installation of the python tree environment ete2: http://etetoolkit.org/. Selenoprofiles_tree_drawer can generate images like those below.
Default mode:
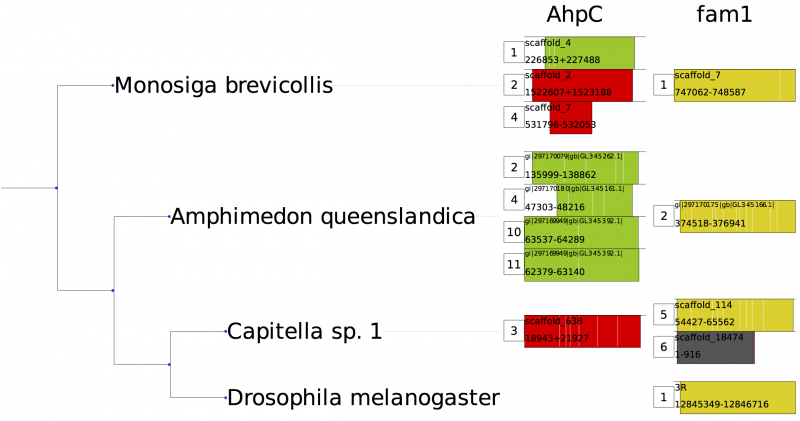
Abstract mode (-a):

Contact
If you need help with selenoprofiles, do not hesitate to contact me by email: marco.mariotti at crg.eu
 Roderic Guigo Lab
Roderic Guigo Lab