There is an increasing
amount of information available in databases that can be
useful to unveil important biological facts.
One of the problems in Bioinformatics is how to structure and store
this data in such a way that it can be readily used for answering
biologically relevant questions.
We will explore one of the methods used to make the information closer
to the questions posed by the biologists.
We know now how to find information about our favourite gene, like
the number of exons, transcripts, the functional domains, etc. We
also know how to view some of the properties of the genome in a region
of interest, e.g.: sequence conservation with other genomes, synteny,
etc. However, can we somehow link all this to ask questions like:
Give me all genes in human with a given Pfam domain and an ortholog gene in mouse and rat, with a sequence similarity greater than 70%.
We will use the Ensmart browser:
Biological databases are usually designed such that
the storage and update of information is optimized, and have
structures (tables, files ) that are very specific to the data at hand.
Complex queries are therefore computationally
expensive as they require a large amount of analyses and computations.
This also often requires specialized software (e.g. Ensembl and UCSC browsers).
Ensmart provides a way of organizing the data such that it is optimized for querying.
The computations are still necessary, but the results are stored in such a way, that
we are able to obtain a very fast answer to queries that involve a non-direct
relationships between attributes. Ensmart is built with pre-computed data and it is
in the way the results are stored that we gain usability.
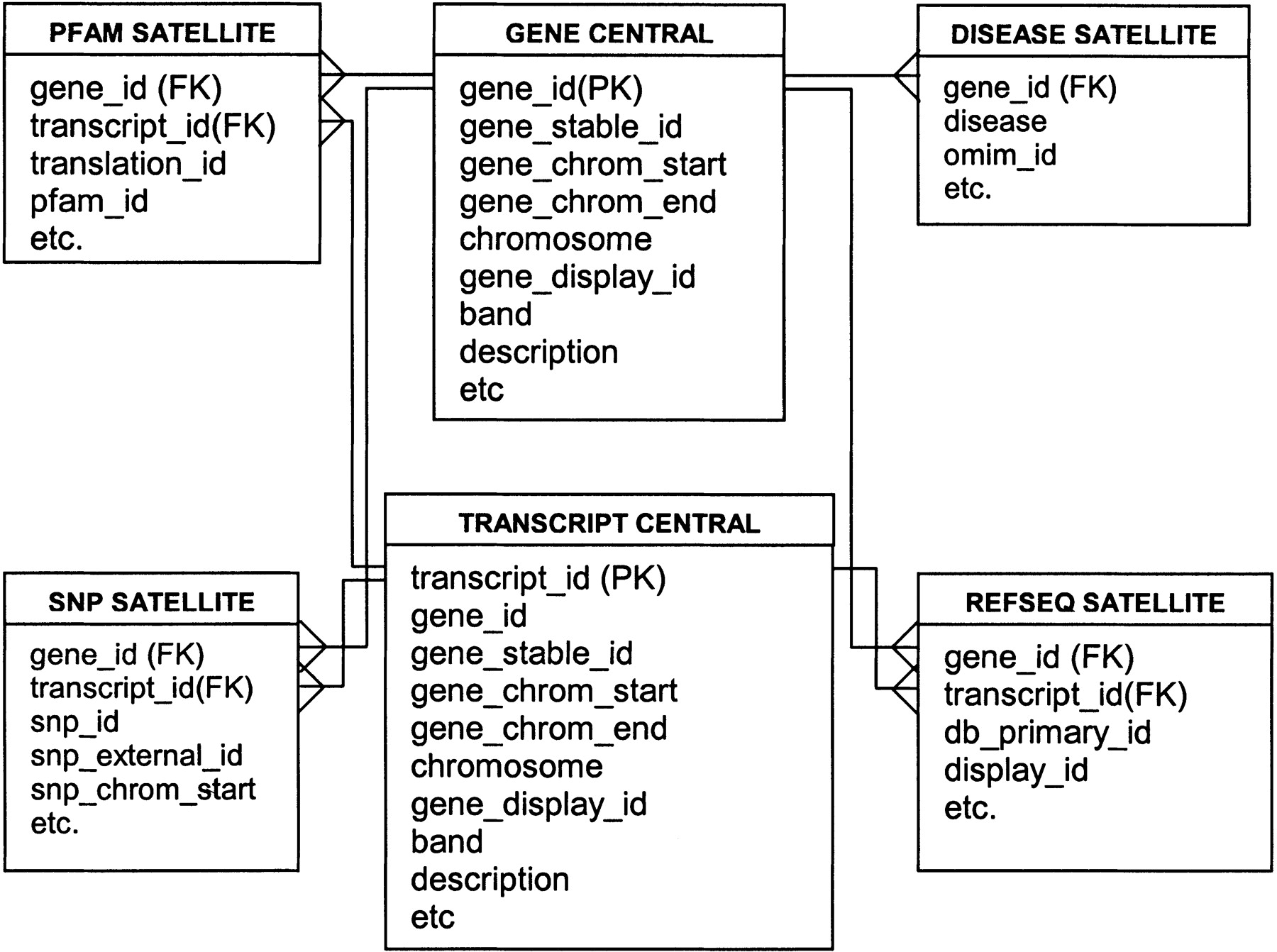
The Ensmart database structure is adapted from a star schema.
There are one or more central tables (gene, transcript, SNP) and these
are linked to a number of satellite tables containing the attributes.
The central table is the source for the constraints and the satellite
is the source for the attributes:

You can read more about it here:
http://www.genome.org/cgi/content/full/14/1/160
We are going to illustrate the use of Ensmart with an example.
We have seen previously (see previous section) that
the ADH gene contains a domain from the Aldo/Keto reductase family with Pfam ID PF00248.
Let's imagine that we are interested in promoters and we want to study the upstream region of human genes with this domain. Additionally we would like to concentrate on genes that have an ortholog in mouse. However, we do not want all of them, we only want to look at those that have non-synonymous SNPs in the coding region of the gene.
We would like to obtain two pieces of information for these genes:
Ensmart is based on a focus, which is the entity about which we ask the questions, and a series of satellites sets of data, which hold the attributes and which are used to do to operations:
Open the browser: www.ensembl.org/EnsMart
How many databases can you see?
How many species can we choose from? Is there anything different in 'species' with respect to the species you can see in www.ensembl.org?
Select dataset: human. Note the versions: NCBI35.
Once we have selected the database, we need to specify the filters. A wide range of filters
can be applied in any combination.
Question: Explore the possible filters
Question: Select the filters according to what we want to answer:
Once we have applied the filter, we can choose which output to generate.
Press the 'next' button. Note the update on the right-hand side.
Question: Explore the four possible types of 'output':
Select the 'sequence' output. Choose 5' end sequence.
Question: Why can we choose between gene and transcript to select the sequences?
Select the upstream region of the gene. You should obtain an output like this:
>ENSG00000165568.4 assembly=NCBI34|chr=10|strand=forward|bases 4821426 to 4822425|region upstream of gene only CTCCCCTGATGGCCAGCACTGAAGACCCAGGCAAGGAACCTAGAAACAAAGCCCTCATCTGGGTGTGGGTGTCCTCAAGG CAGTAGGACTCCCAGGGCTGAGGGGGGCAATGAAGGGGGAGCTGTAAGCTCCAGGAGAGATAAGAGGGGCGTCGGAAGGC TCCCTTGACCCCTCTTTCCCTCCACTGGCCCTGGGGGAGCCCAGTCCACTCATAAGGGGGGTGTCCAGTCCACCCCATCC ...
We can go back in the browser to select the expression patter of these genes.
In the Features section select one of the sources of expression.
Question:
Carry out the following searches:
Other examples of data you can retrieve with Ensmart:
As the idea behind Ensmart is data-driven rather than data-dependent,
the same approach is extensible to other
data sources. This is achieved by collecting the raw data from a database and generating the
tables in a star schema. Once the satellite tables are defined the structure and dependencies can
be automatically generated. Any additional domain specific information can be added in the form of external
lookup tables.
Go to the web page of Biomart
We can see that so far it has been applied to other databases:
This picture illustrates what ESTGenes are:
Click on the BioMart logo.
Hint: Obtain first from Uniprot the "Uniprot Accession Identifiers" of those
proteins with an associated PDB ID. Then search for the entries in the MSD database
corresponding to these accession IDs.
Questions:
Some take-home messages: