Gene Prediction
Written by Enrique Blanco
Overview
In this section we use several gene prediction programs on a particular
genomic DNA sequence. For each of these programs we obtain a prediction of
a candidate gene and we will analyze the differences between predictions and
the annotation of the real gene.
The programs we are going to use are geneid, genscan and
fgenesh, which are available through a web interface. In these, and in
many other tools in the web, we access a form where we can paste, or
submit, the sequence we want to analyze, and then we press a button in
the form that starts the computing process in some computer where the program
runs. Once this process is finished, we get a new page in
our browser with the results, which in this case should be a predicted gene.
A Genomic DNA Sequence
We are going to work with the sequence HS307871, which
is stored in FASTA format. This sequence contains one gene, annotated in the
following EMBL and
NCBI records. Try to identify in these
records the different pieces of information related to the annotation of the gene.
geneid
In order to use geneid follow these steps:
- Connect to the geneid server by following
this link.
- Paste the DNA sequence.
- Select organism (human).
- Run geneid with different (output) parameters:
- Searching signals: Select acceptors, donors, start and stop codons. For each type of signal, try to find the real ones.
- Searching exons: Select All exons and try to find the real ones.
- Finding genes: You do not need to select any option (default behavior).
- Compare the prediction with the real annotation.
- By taking a look to the graphical representation of the predicted sites and
exons.
- By inspection of the output and the EMBL/NCBI record.
- By taking a look to the graphical representation of both, the output and the EMBL/NCBI annotation
in this link.
- Improve the prediction from some confirmed evidence.
- Below the text box where we pasted the DNA sequence, we can find a text box where we can paste
evidences, which should consist of one or more exons (in GFF format) that are, e.g.,
experimentally confirmed.
- In this case we are going to paste as evidence the first exon which has not been predicted.
Select and copy the GFF line corresponding to this exon contained in this file.
- Paste the line into the evidences text box, and run again geneid on the sequence.
- Compare the result with the real annotation. What has changed from the previous prediction?
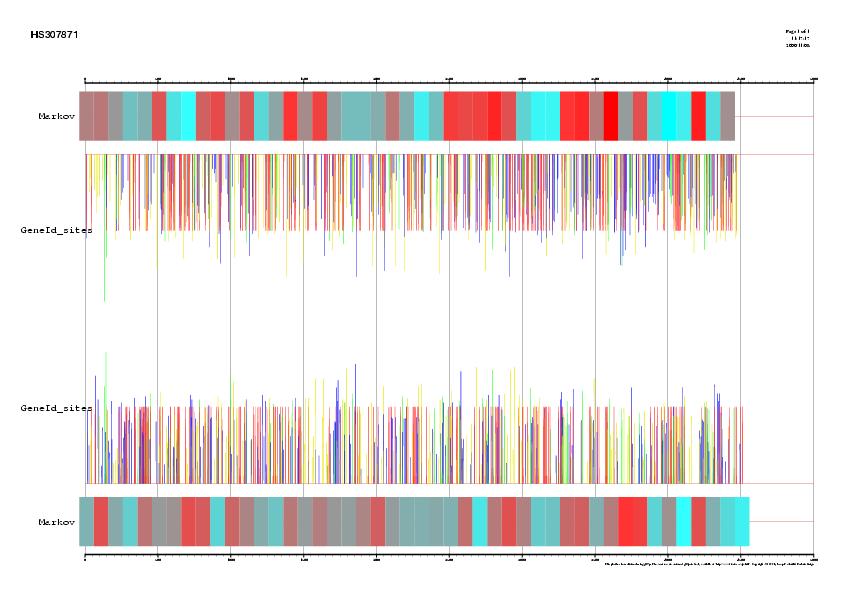 |
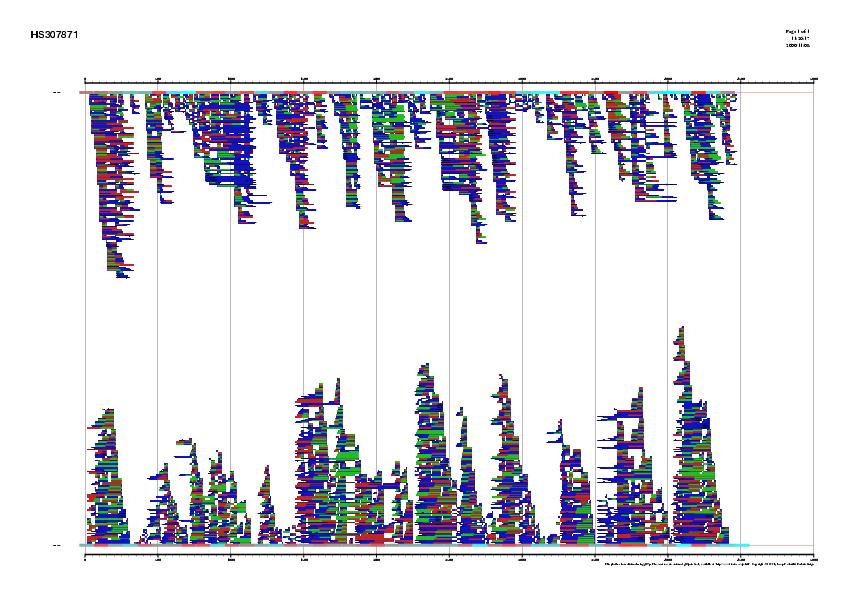 |
 |
|
Signal, exons and genes predicted by geneid in the sequence HS307871
|
genscan
In order to use genscan follow these steps:
- Connect to the genscan server by following
this link.
- Paste the DNA sequence.
- Select organism (vertebrate).
- Compare the prediction with the real annotation.
- By inspection of the output and the EMBL/NCBI record.
- By taking a look to the graphical representation of both, the output and the EMBL/NCBI annotation
in this link.
fgenesh
In order to use fgenesh follow these steps:
- Connect to the fgenesh server by following
this link.
- Paste the DNA sequence.
- Select organism (human).
- Compare the prediction with the real annotation.
- By inspection of the output and the EMBL/NCBI record.
- By taking a look to the graphical representation of both, the output and the EMBL/NCBI annotation
in this link.
Current Annotations in the Genomic DNA Sequence
We can see the annotation of the gene together with the three predicted genes by geneid,
genscan and fgenesh by following this link.
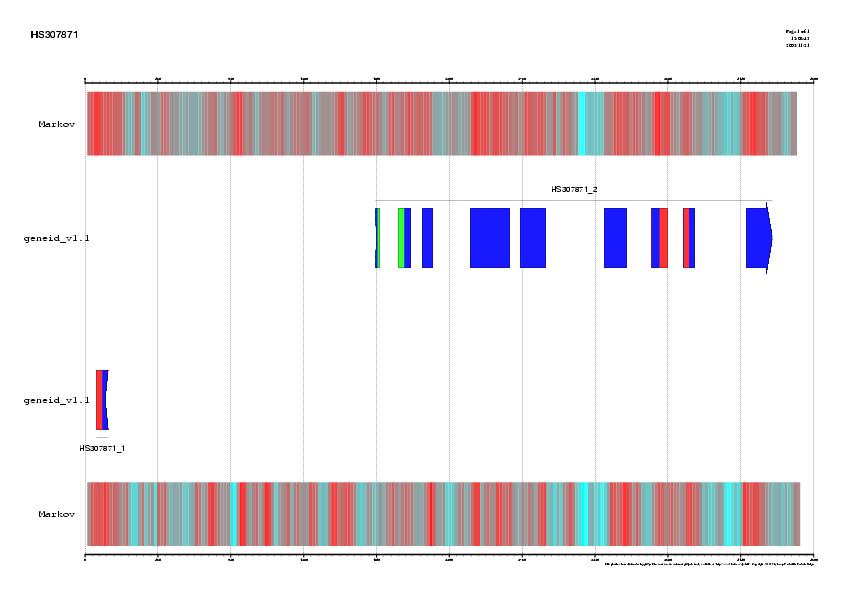
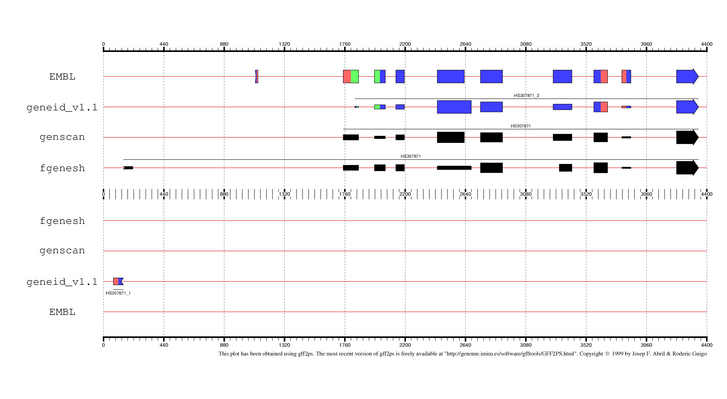 |
|
EMBL annotation and genes predicted by
geneid, GENSCAN and FGENESH in the sequence HS307871
|
Go to the page where we saw the NCBI record, click on the link CDS, and,
next to the Display button, unroll the menu box and select the display
option FASTA. Now press the button Display, and we will obtain the
protein-coding DNA sequence of this gene in FASTA format.
Select the entire sequence (first line is not necessary) and go to the UCSC genome
BLAT search by following this link.
In the big text box, paste the coding sequence we just copied, and press the Submit
button on the top-right corner of this page.
The result is a single match, where we find two links, browser and
details. Visit first the details link and try to understand the
the information provided there. Then go backwards and visit the browser
link where we will see where this gene is located within the Human genome, as well
as other annotated information as EST spliced alignments, etc.
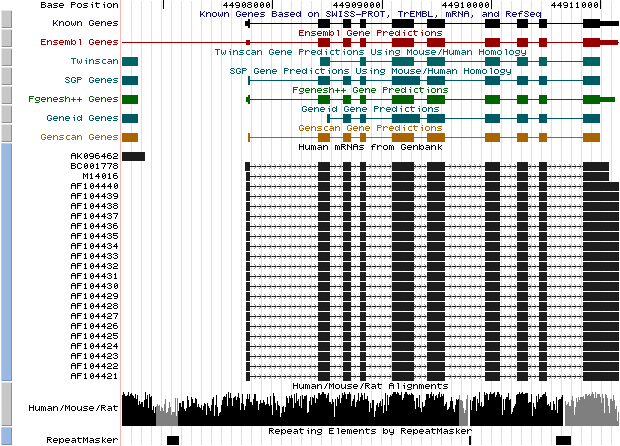 |
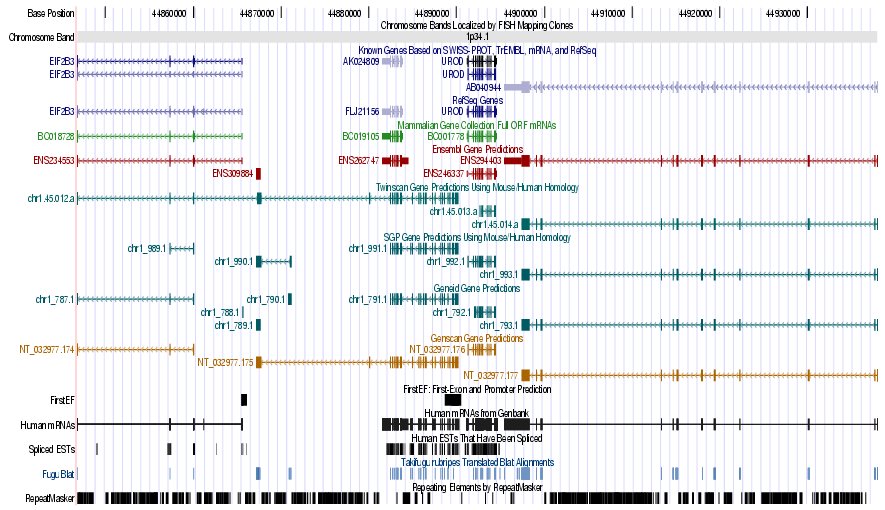 |
Left) UCSC genome browser representation of the region
containing the gene uroporphyrinogen decarboxylase (URO-D)
Right) UCSC genome browser representation of the contex (100Kbps) region around
the gene uroporphyrinogen decarboxylase (URO-D).
|


